Run MBNMA time-course models
mb.run.RdFits a Bayesian time-course model for model-based network meta-analysis (MBNMA) that can account for repeated measures over time within studies by applying a desired time-course function. Follows the methods of pedder2019;textualMBNMAtime.
Usage
mb.run(
network,
fun = tpoly(degree = 1),
positive.scale = FALSE,
intercept = NULL,
link = "identity",
sdscale = FALSE,
parameters.to.save = NULL,
rho = 0,
covar = "varadj",
corparam = FALSE,
class.effect = list(),
UME = FALSE,
parallel = FALSE,
priors = NULL,
n.iter = 20000,
n.chains = 3,
n.burnin = floor(n.iter/2),
n.thin = max(1, floor((n.iter - n.burnin)/1000)),
pD = TRUE,
model.file = NULL,
jagsdata = NULL,
...
)Arguments
- network
An object of class
"mb.network".- fun
An object of class
"timefun"generated (see Details) using any oftloglin(),tpoly(),titp(),temax(),tfpoly(),tspline()ortuser()- positive.scale
A boolean object that indicates whether all continuous mean responses (y) are positive and therefore whether the baseline response should be given a prior that constrains it to be positive (e.g. for scales that cannot be <0).
- intercept
A boolean object that indicates whether an intercept (written as
alphain the model) is to be included. If left asNULL(the default), an intercept will be included only for studies reporting absolute means, and will be excluded for studies reporting change from baseline (as indicated innetwork$cfb).- link
Can take either
"identity"(the default),"log"(for modelling Ratios of Means friedrich2011MBNMAtime) or"smd"(for modelling Standardised Mean Differences - although this also corresponds to an identity link function).- sdscale
Logical object to indicate whether to write a model that specifies a reference SD for standardising when modelling using Standardised Mean Differences. Specifying
sdscale=TRUEwill therefore only modify the model if link function is set to SMD (link="smd").- parameters.to.save
A character vector containing names of parameters to monitor in JAGS
- rho
The correlation coefficient when modelling within-study correlation between time points. The default is a string representing a prior distribution in JAGS, indicating that it be estimated from the data (e.g.
rho="dunif(0,1)").rhoalso be assigned a numeric value (e.g.rho=0.7), which fixesrhoin the model to this value (e.g. for use in a deterministic sensitivity analysis). If set torho=0(the default) then this implies modelling no correlation between time points.- covar
A character specifying the covariance structure to use for modelling within-study correlation between time-points. This can be done by specifying one of the following:
"varadj"- a univariate likelihood with a variance adjustment to assume a constant correlation between subsequent time points jansen2015MBNMAtime. This is the default."CS"- a multivariate normal likelihood with a compound symmetry structure"AR1"- a multivariate normal likelihood with an autoregressive AR1 structure
- corparam
A boolean object that indicates whether correlation should be modeled between relative effect time-course parameters. Default is
FALSEand this is automatically set toFALSEif class effects are modeled. Setting it toTRUEmodels correlation between time-course parameters. This can help identify parameters that are estimated poorly for some treatments by allowing sharing of information between parameters for different treatments in the network, but may also cause some shrinkage.- class.effect
A list of named strings that determines which time-course parameters to model with a class effect and what that effect should be (
"common"or"random"). For example:list(emax="common", et50="random").- UME
Can take either
TRUEorFALSE(for an unrelated mean effects model on all or no time-course parameters respectively) or can be a vector of parameter name strings to model as UME. For example:c("beta.1", "beta.2").- parallel
A boolean value that indicates whether JAGS should be run in parallel (
TRUE) or not (FALSE). IfTRUEthen the number of cores to use is automatically calculated. Functions that involve updating the model (e.g.devplot(),fitplot()) cannot be used with models implemented in parallel.- priors
A named list of parameter values (without indices) and replacement prior distribution values given as strings using distributions as specified in JAGS syntax (see jagsmanual;textualMBNMAtime).
- n.iter
number of total iterations per chain (including burn in; default: 20000)
- n.chains
number of Markov chains (default: 3)
- n.burnin
length of burn in, i.e. number of iterations to discard at the beginning. Default is
n.iter/2``, that is, discarding the first half of the simulations. Ifn.burnin` is 0, jags() will run 100 iterations for adaption.- n.thin
thinning rate. Must be a positive integer. Set
n.thin > 1`` to save memory and computation time ifn.iteris large. Default ismax(1, floor(n.chains * (n.iter-n.burnin) / 1000))“ which will only thin if there are at least 2000 simulations.- pD
logical; if
TRUE(the default) then adds the computation of pD, using the method of plummer2008MBNMAtime. IfFALSEthen uses the approximation ofpD=var(deviance) / 2(often referred to as pV).- model.file
The file path to a JAGS model (.jags file) that can be used to overwrite the JAGS model that is automatically written based on the specified options in
MBNMAtime. Useful for adding further model flexibility.- jagsdata
A named list of the data objects to be used in the JAGS model. Only required if users are defining their own JAGS model using
model.file. Format should match that of standard models fitted inMBNMAtime(seembnma$model.arg$jagsdata)- ...
Arguments to be sent to R2jags.
Value
An object of S3 class c("mbnma", "rjags")`` containing parameter results from the model. Can be summarized by print()and can check traceplots usingR2jags::traceplot()or various functions from the packagemcmcplots`.#'
If there are errors in the JAGS model code then the object will be a list
consisting of two elements - an error message from JAGS that can help with
debugging and model.arg, a list of arguments provided to mb.run()
which includes jagscode, the JAGS code for the model that can help
users identify the source of the error.
Time-course parameters
Nodes that are automatically monitored (if present in the model) have the
same name as in the time-course function for named time-course parameters (e.g. emax).
However, for named only as beta.1, beta.2, beta.3 or beta.4 parameters
may have an alternative interpretation.
Details of the interpretation and model specification of different parameters can be shown by using the
summary() method on an "mbnma" object generated by mb.run().
Parameters modelled using relative effects
If pooling is relative (e.g.
pool.1="rel") for a given parameter then the named parameter (e.g.emax) or a numbereddparameter (e.g.d.1) corresponds to the pooled relative effect for a given treatment compared to the network reference treatment for this time-course parameter.sd.followed by a named (e.g.emax,beta.1) is the between-study SD (heterogeneity) for relative effects, reported if pooling for a time-course parameter is relative (e.g.pool.1="rel") and the method for synthesis is random (e.g.method.1="random).If class effects are modelled, parameters for classes are represented by the upper case name of the time-course parameter they correspond to. For example if
class.effect=list(emax="random"), relative class effects will be represented byEMAX. The SD of the class effect (e.g.sd.EMAX,sd.BETA.1) is the SD of treatments within a class for the time-course parameter they correspond to.
Parameters modelled using absolute effects
If pooling is absolute (e.g.
pool.1="abs") for a given parameter then the named parameter (e.g.emax) or a numberedbetaparameter (e.g.beta.1) corresponds to the estimated absolute effect for this time-course parameter.For an absolute time-course parameter if the corresponding method is common (e.g.
method.1="common") the parameter corresponds to a single common parameter estimated across all studies and treatments. If the corresponding method is random (e.g.method.1="random") then parameter is a mean effect around which the study-level absolute effects vary with SD corresponding tosd.followed by the named parameter (e.g.sd.emax,sd.beta.1) .
Other model parameters
rhoThe correlation coefficient for correlation between time-points. Its interpretation will differ depending on the covariance structure specified incovartotresdevThe residual deviance of the modeldevianceThe deviance of the model
Time-course function
Several general time-course functions with up to 4 time-course parameters are provided, but a user-defined time-course relationship can instead be used. Details can be found in the respective help files for each function.
Available time-course functions are:
Correlation between observations
When modelling correlation between observations using rho, values for rho must imply a
positive semidefinite covariance matrix.
Advanced options
model.file and jagsdata can be used to run an edited JAGS model and dataset. This allows
users considerably more modelling flexibility than is possible using the basic MBNMAtime syntax,
though requires strong understanding of JAGS and the MBNMA modelling framework. Treatment-specific
priors, meta-regression and bias-adjustment are all possible in this way, and it allows users to
make use of the subsequent functions in MBNMAtime (plotting, prediction, ranking) whilst fitting
these more complex models.
Examples
# \donttest{
# Create mb.network object
network <- mb.network(osteopain)
#> Reference treatment is `Pl_0`
#> Studies reporting change from baseline automatically identified from the data
# Fit a linear time-course MBNMA with:
# random relative treatment effects on the slope
mb.run(network, fun=tpoly(degree=1, pool.1="rel", method.1="random"))
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 417
#> Unobserved stochastic nodes: 163
#> Total graph size: 7701
#>
#> Initializing model
#>
#> Inference for Bugs model at "/tmp/RtmpPWTcs6/file1eda782fc3e5", fit using jags,
#> 3 chains, each with 20000 iterations (first 10000 discarded), n.thin = 10
#> n.sims = 3000 iterations saved. Running time = 7.582 secs
#> mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat
#> d.1[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000
#> d.1[2] -0.064 0.046 -0.155 -0.094 -0.063 -0.035 0.024 1.002
#> d.1[3] -0.094 0.018 -0.129 -0.106 -0.094 -0.082 -0.058 1.001
#> d.1[4] -0.094 0.044 -0.180 -0.123 -0.093 -0.065 -0.009 1.001
#> d.1[5] -0.043 0.053 -0.147 -0.079 -0.043 -0.008 0.062 1.002
#> d.1[6] 0.002 0.070 -0.138 -0.045 0.002 0.050 0.142 1.001
#> d.1[7] -0.148 0.035 -0.218 -0.171 -0.148 -0.125 -0.083 1.001
#> d.1[8] -0.028 0.069 -0.169 -0.072 -0.029 0.016 0.106 1.002
#> d.1[9] -0.290 0.049 -0.389 -0.322 -0.289 -0.257 -0.196 1.001
#> d.1[10] -0.306 0.070 -0.445 -0.353 -0.305 -0.258 -0.169 1.001
#> d.1[11] -0.076 0.038 -0.152 -0.102 -0.075 -0.051 -0.001 1.001
#> d.1[12] -0.069 0.037 -0.142 -0.093 -0.069 -0.044 0.004 1.002
#> d.1[13] -0.084 0.046 -0.173 -0.116 -0.083 -0.052 0.007 1.001
#> d.1[14] -0.082 0.063 -0.208 -0.123 -0.083 -0.041 0.043 1.001
#> d.1[15] -0.128 0.025 -0.178 -0.145 -0.128 -0.111 -0.078 1.001
#> d.1[16] -0.135 0.039 -0.211 -0.162 -0.135 -0.109 -0.058 1.001
#> d.1[17] 0.112 0.066 -0.024 0.069 0.113 0.155 0.244 1.002
#> d.1[18] -0.128 0.049 -0.221 -0.159 -0.128 -0.097 -0.031 1.001
#> d.1[19] -0.123 0.077 -0.277 -0.173 -0.124 -0.073 0.034 1.001
#> d.1[20] -0.115 0.048 -0.213 -0.145 -0.114 -0.083 -0.021 1.001
#> d.1[21] -0.426 0.079 -0.580 -0.478 -0.429 -0.373 -0.273 1.001
#> d.1[22] -0.171 0.042 -0.256 -0.199 -0.170 -0.144 -0.088 1.001
#> d.1[23] -0.031 0.041 -0.113 -0.058 -0.031 -0.004 0.049 1.001
#> d.1[24] -0.040 0.042 -0.120 -0.068 -0.041 -0.012 0.042 1.002
#> d.1[25] -0.067 0.042 -0.150 -0.093 -0.066 -0.039 0.015 1.001
#> d.1[26] -0.085 0.063 -0.209 -0.125 -0.085 -0.044 0.037 1.001
#> d.1[27] -0.067 0.066 -0.196 -0.112 -0.067 -0.024 0.062 1.001
#> d.1[28] -0.094 0.065 -0.220 -0.138 -0.094 -0.049 0.035 1.001
#> d.1[29] -0.078 0.066 -0.210 -0.123 -0.078 -0.034 0.054 1.001
#> rho 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000
#> sd.beta.1 0.073 0.009 0.056 0.066 0.072 0.079 0.094 1.001
#> totresdev 9298.979 16.866 9267.942 9287.281 9298.448 9309.983 9333.912 1.001
#> deviance 8418.940 16.866 8387.903 8407.242 8418.408 8429.944 8453.873 1.001
#> n.eff
#> d.1[1] 1
#> d.1[2] 1600
#> d.1[3] 3000
#> d.1[4] 3000
#> d.1[5] 1600
#> d.1[6] 3000
#> d.1[7] 3000
#> d.1[8] 1200
#> d.1[9] 3000
#> d.1[10] 3000
#> d.1[11] 2000
#> d.1[12] 3000
#> d.1[13] 3000
#> d.1[14] 2300
#> d.1[15] 3000
#> d.1[16] 3000
#> d.1[17] 1900
#> d.1[18] 2500
#> d.1[19] 2500
#> d.1[20] 3000
#> d.1[21] 3000
#> d.1[22] 3000
#> d.1[23] 3000
#> d.1[24] 2400
#> d.1[25] 3000
#> d.1[26] 3000
#> d.1[27] 2100
#> d.1[28] 3000
#> d.1[29] 3000
#> rho 1
#> sd.beta.1 3000
#> totresdev 2100
#> deviance 2100
#>
#> For each parameter, n.eff is a crude measure of effective sample size,
#> and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
#>
#> DIC info (using the rule: pV = var(deviance)/2)
#> pV = 142.2 and DIC = 8549.8
#> DIC info (using the rule: pD = Dbar-Dhat, computed via 'rjags::dic.samples')
#> pD = 131.4 and DIC = 8549.7
#> DIC is an estimate of expected predictive error (lower deviance is better).
# Fit an emax time-course MBNMA with:
# fixed relative treatment effects on emax
# a common parameter estimated independently of treatment
# a common Hill parameter estimated independently of treatment
# a prior for the Hill parameter (normal with mean 0 and precision 0.1)
# data reported as change from baseline
result <- mb.run(network, fun=temax(pool.emax="rel", method.emax="common",
pool.et50="abs", method.et50="common",
pool.hill="abs", method.hill="common"),
priors=list(hill="dunif(0.5, 2)"),
intercept=TRUE)
#> 'et50' parameters must take positive values.
#> Default half-normal prior restricts posterior to positive values.
#> 'hill' parameters must take positive values.
#> Default half-normal prior restricts posterior to positive values.
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 417
#> Unobserved stochastic nodes: 90
#> Total graph size: 7740
#>
#> Initializing model
#>
#### commented out to prevent errors from JAGS version in github actions build ####
# Fit a log-linear MBNMA with:
# random relative treatment effects on the rate
# an autoregressive AR1 covariance structure
# modelled as standardised mean differences
# copdnet <- mb.network(copd)
# result <- mb.run(copdnet, fun=tloglin(pool.rate="rel", method.rate="random"),
# covar="AR1", rho="dunif(0,1)", link="smd")
####### Examine MCMC diagnostics (using mcmcplots package) #######
# Traceplots
# mcmcplots::traplot(result)
# Plots for assessing convergence
# mcmcplots::mcmcplot(result, c("rate", "sd.rate", "rho"))
########## Output ###########
# Print R2jags output and summary
print(result)
#> Inference for Bugs model at "/tmp/RtmpPWTcs6/file1eda455b940b", fit using jags,
#> 3 chains, each with 20000 iterations (first 10000 discarded), n.thin = 10
#> n.sims = 3000 iterations saved. Running time = 44.971 secs
#> mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
#> emax[1] 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1
#> emax[2] -0.643 0.106 -0.851 -0.714 -0.642 -0.571 -0.432 1.001 3000
#> emax[3] -0.934 0.044 -1.017 -0.964 -0.935 -0.905 -0.847 1.002 1200
#> emax[4] -1.043 0.104 -1.250 -1.111 -1.043 -0.971 -0.843 1.001 3000
#> emax[5] -0.640 0.198 -1.031 -0.770 -0.637 -0.512 -0.248 1.001 3000
#> emax[6] -0.112 0.141 -0.394 -0.211 -0.112 -0.016 0.162 1.001 3000
#> emax[7] -1.231 0.070 -1.370 -1.275 -1.230 -1.184 -1.095 1.002 1100
#> emax[8] -0.115 0.143 -0.397 -0.211 -0.115 -0.022 0.179 1.001 3000
#> emax[9] -1.896 0.108 -2.111 -1.969 -1.895 -1.823 -1.683 1.001 3000
#> emax[10] -1.896 0.156 -2.207 -2.000 -1.897 -1.791 -1.601 1.002 1200
#> emax[11] -0.878 0.061 -0.998 -0.920 -0.878 -0.837 -0.760 1.001 3000
#> emax[12] -0.861 0.063 -0.983 -0.903 -0.860 -0.818 -0.735 1.001 3000
#> emax[13] -1.064 0.076 -1.216 -1.116 -1.063 -1.013 -0.917 1.001 3000
#> emax[14] -0.996 0.124 -1.244 -1.079 -0.994 -0.914 -0.755 1.002 1000
#> emax[15] -1.318 0.064 -1.443 -1.362 -1.320 -1.275 -1.190 1.003 1400
#> emax[16] -1.224 0.074 -1.369 -1.275 -1.223 -1.172 -1.080 1.004 520
#> emax[17] 0.359 0.138 0.088 0.266 0.355 0.452 0.633 1.001 3000
#> emax[18] -0.927 0.088 -1.097 -0.986 -0.927 -0.866 -0.760 1.004 530
#> emax[19] -1.287 0.237 -1.763 -1.443 -1.286 -1.125 -0.825 1.005 470
#> emax[20] -0.882 0.101 -1.084 -0.949 -0.882 -0.814 -0.686 1.001 2800
#> emax[21] -2.641 0.205 -3.036 -2.779 -2.638 -2.503 -2.248 1.002 1600
#> emax[22] -1.311 0.114 -1.541 -1.393 -1.313 -1.231 -1.091 1.002 1300
#> emax[23] -0.213 0.069 -0.349 -0.261 -0.213 -0.167 -0.079 1.001 3000
#> emax[24] -0.332 0.070 -0.473 -0.378 -0.329 -0.285 -0.199 1.002 1400
#> emax[25] -0.766 0.071 -0.905 -0.813 -0.767 -0.717 -0.629 1.004 590
#> emax[26] -0.830 0.096 -1.020 -0.894 -0.831 -0.764 -0.647 1.002 1900
#> emax[27] -0.787 0.120 -1.024 -0.868 -0.785 -0.707 -0.549 1.002 1600
#> emax[28] -1.025 0.123 -1.264 -1.111 -1.022 -0.938 -0.789 1.001 3000
#> emax[29] -0.832 0.120 -1.070 -0.912 -0.833 -0.751 -0.594 1.001 3000
#> et50 0.462 0.044 0.383 0.431 0.459 0.490 0.556 1.004 660
#> hill 0.578 0.069 0.502 0.524 0.558 0.615 0.756 1.009 470
#> rho 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 1
#> totresdev 837.536 13.491 813.247 828.124 836.913 846.042 865.878 1.001 3000
#> deviance -42.504 13.491 -66.793 -51.916 -43.126 -33.997 -14.161 1.001 3000
#>
#> For each parameter, n.eff is a crude measure of effective sample size,
#> and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
#>
#> DIC info (using the rule: pV = var(deviance)/2)
#> pV = 91.0 and DIC = 46.6
#> DIC info (using the rule: pD = Dbar-Dhat, computed via 'rjags::dic.samples')
#> pD = 89.8 and DIC = 47.1
#> DIC is an estimate of expected predictive error (lower deviance is better).
summary(result)
#> ========================================
#> Time-course MBNMA
#> ========================================
#>
#> Time-course function: emax
#> Data modelled with intercept
#>
#> emax parameter
#> Pooling: relative effects
#> Method: common treatment effects
#>
#> |Treatment |Parameter | Median| 2.5%| 97.5%|
#> |:---------|:---------|-------:|-------:|-------:|
#> |Pl_0 |emax[1] | 0.0000| 0.0000| 0.0000|
#> |Ce_100 |emax[2] | -0.6423| -0.8510| -0.4317|
#> |Ce_200 |emax[3] | -0.9351| -1.0171| -0.8471|
#> |Ce_400 |emax[4] | -1.0429| -1.2504| -0.8425|
#> |Du_90 |emax[5] | -0.6375| -1.0310| -0.2484|
#> |Et_10 |emax[6] | -0.1124| -0.3940| 0.1615|
#> |Et_30 |emax[7] | -1.2301| -1.3700| -1.0952|
#> |Et_5 |emax[8] | -0.1153| -0.3968| 0.1785|
#> |Et_60 |emax[9] | -1.8947| -2.1109| -1.6828|
#> |Et_90 |emax[10] | -1.8967| -2.2075| -1.6006|
#> |Lu_100 |emax[11] | -0.8781| -0.9982| -0.7599|
#> |Lu_200 |emax[12] | -0.8600| -0.9828| -0.7351|
#> |Lu_400 |emax[13] | -1.0630| -1.2162| -0.9168|
#> |Lu_NA |emax[14] | -0.9941| -1.2437| -0.7549|
#> |Na_1000 |emax[15] | -1.3197| -1.4433| -1.1901|
#> |Na_1500 |emax[16] | -1.2232| -1.3686| -1.0799|
#> |Na_250 |emax[17] | 0.3545| 0.0880| 0.6334|
#> |Na_750 |emax[18] | -0.9272| -1.0971| -0.7603|
#> |Ox_44 |emax[19] | -1.2864| -1.7631| -0.8246|
#> |Ro_12 |emax[20] | -0.8819| -1.0838| -0.6863|
#> |Ro_125 |emax[21] | -2.6379| -3.0358| -2.2477|
#> |Ro_25 |emax[22] | -1.3126| -1.5407| -1.0911|
#> |Tr_100 |emax[23] | -0.2135| -0.3493| -0.0794|
#> |Tr_200 |emax[24] | -0.3287| -0.4733| -0.1993|
#> |Tr_300 |emax[25] | -0.7665| -0.9052| -0.6289|
#> |Tr_400 |emax[26] | -0.8315| -1.0199| -0.6468|
#> |Va_10 |emax[27] | -0.7852| -1.0240| -0.5488|
#> |Va_20 |emax[28] | -1.0217| -1.2643| -0.7894|
#> |Va_5 |emax[29] | -0.8325| -1.0705| -0.5943|
#>
#>
#> et50 parameter
#> Pooling: absolute effects
#> Method: common treatment effects
#>
#> |Treatment |Parameter | Median| 2.5%| 97.5%|
#> |:---------|:---------|------:|------:|------:|
#> |Pl_0 |et50 | 0.4593| 0.3834| 0.5561|
#> |Ce_100 |et50 | 0.4593| 0.3834| 0.5561|
#> |Ce_200 |et50 | 0.4593| 0.3834| 0.5561|
#> |Ce_400 |et50 | 0.4593| 0.3834| 0.5561|
#> |Du_90 |et50 | 0.4593| 0.3834| 0.5561|
#> |Et_10 |et50 | 0.4593| 0.3834| 0.5561|
#> |Et_30 |et50 | 0.4593| 0.3834| 0.5561|
#> |Et_5 |et50 | 0.4593| 0.3834| 0.5561|
#> |Et_60 |et50 | 0.4593| 0.3834| 0.5561|
#> |Et_90 |et50 | 0.4593| 0.3834| 0.5561|
#> |Lu_100 |et50 | 0.4593| 0.3834| 0.5561|
#> |Lu_200 |et50 | 0.4593| 0.3834| 0.5561|
#> |Lu_400 |et50 | 0.4593| 0.3834| 0.5561|
#> |Lu_NA |et50 | 0.4593| 0.3834| 0.5561|
#> |Na_1000 |et50 | 0.4593| 0.3834| 0.5561|
#> |Na_1500 |et50 | 0.4593| 0.3834| 0.5561|
#> |Na_250 |et50 | 0.4593| 0.3834| 0.5561|
#> |Na_750 |et50 | 0.4593| 0.3834| 0.5561|
#> |Ox_44 |et50 | 0.4593| 0.3834| 0.5561|
#> |Ro_12 |et50 | 0.4593| 0.3834| 0.5561|
#> |Ro_125 |et50 | 0.4593| 0.3834| 0.5561|
#> |Ro_25 |et50 | 0.4593| 0.3834| 0.5561|
#> |Tr_100 |et50 | 0.4593| 0.3834| 0.5561|
#> |Tr_200 |et50 | 0.4593| 0.3834| 0.5561|
#> |Tr_300 |et50 | 0.4593| 0.3834| 0.5561|
#> |Tr_400 |et50 | 0.4593| 0.3834| 0.5561|
#> |Va_10 |et50 | 0.4593| 0.3834| 0.5561|
#> |Va_20 |et50 | 0.4593| 0.3834| 0.5561|
#> |Va_5 |et50 | 0.4593| 0.3834| 0.5561|
#>
#>
#> hill parameter
#> Pooling: absolute effects
#> Method: common treatment effects
#>
#> |Treatment |Parameter | Median| 2.5%| 97.5%|
#> |:---------|:---------|------:|------:|------:|
#> |Pl_0 |hill | 0.5584| 0.5023| 0.7563|
#> |Ce_100 |hill | 0.5584| 0.5023| 0.7563|
#> |Ce_200 |hill | 0.5584| 0.5023| 0.7563|
#> |Ce_400 |hill | 0.5584| 0.5023| 0.7563|
#> |Du_90 |hill | 0.5584| 0.5023| 0.7563|
#> |Et_10 |hill | 0.5584| 0.5023| 0.7563|
#> |Et_30 |hill | 0.5584| 0.5023| 0.7563|
#> |Et_5 |hill | 0.5584| 0.5023| 0.7563|
#> |Et_60 |hill | 0.5584| 0.5023| 0.7563|
#> |Et_90 |hill | 0.5584| 0.5023| 0.7563|
#> |Lu_100 |hill | 0.5584| 0.5023| 0.7563|
#> |Lu_200 |hill | 0.5584| 0.5023| 0.7563|
#> |Lu_400 |hill | 0.5584| 0.5023| 0.7563|
#> |Lu_NA |hill | 0.5584| 0.5023| 0.7563|
#> |Na_1000 |hill | 0.5584| 0.5023| 0.7563|
#> |Na_1500 |hill | 0.5584| 0.5023| 0.7563|
#> |Na_250 |hill | 0.5584| 0.5023| 0.7563|
#> |Na_750 |hill | 0.5584| 0.5023| 0.7563|
#> |Ox_44 |hill | 0.5584| 0.5023| 0.7563|
#> |Ro_12 |hill | 0.5584| 0.5023| 0.7563|
#> |Ro_125 |hill | 0.5584| 0.5023| 0.7563|
#> |Ro_25 |hill | 0.5584| 0.5023| 0.7563|
#> |Tr_100 |hill | 0.5584| 0.5023| 0.7563|
#> |Tr_200 |hill | 0.5584| 0.5023| 0.7563|
#> |Tr_300 |hill | 0.5584| 0.5023| 0.7563|
#> |Tr_400 |hill | 0.5584| 0.5023| 0.7563|
#> |Va_10 |hill | 0.5584| 0.5023| 0.7563|
#> |Va_20 |hill | 0.5584| 0.5023| 0.7563|
#> |Va_5 |hill | 0.5584| 0.5023| 0.7563|
#>
#>
#>
#> Correlation between time points
#> Covariance structure: varadj
#> Rho assigned a numeric value: 0
#>
#> #### Model Fit Statistics ####
#>
#> Effective number of parameters:
#> pD calculated using the Kullback-Leibler divergence = 90
#> Deviance = -43
#> Residual deviance = 837
#> Deviance Information Criterion (DIC) = 47
#>
# Plot forest plot of results
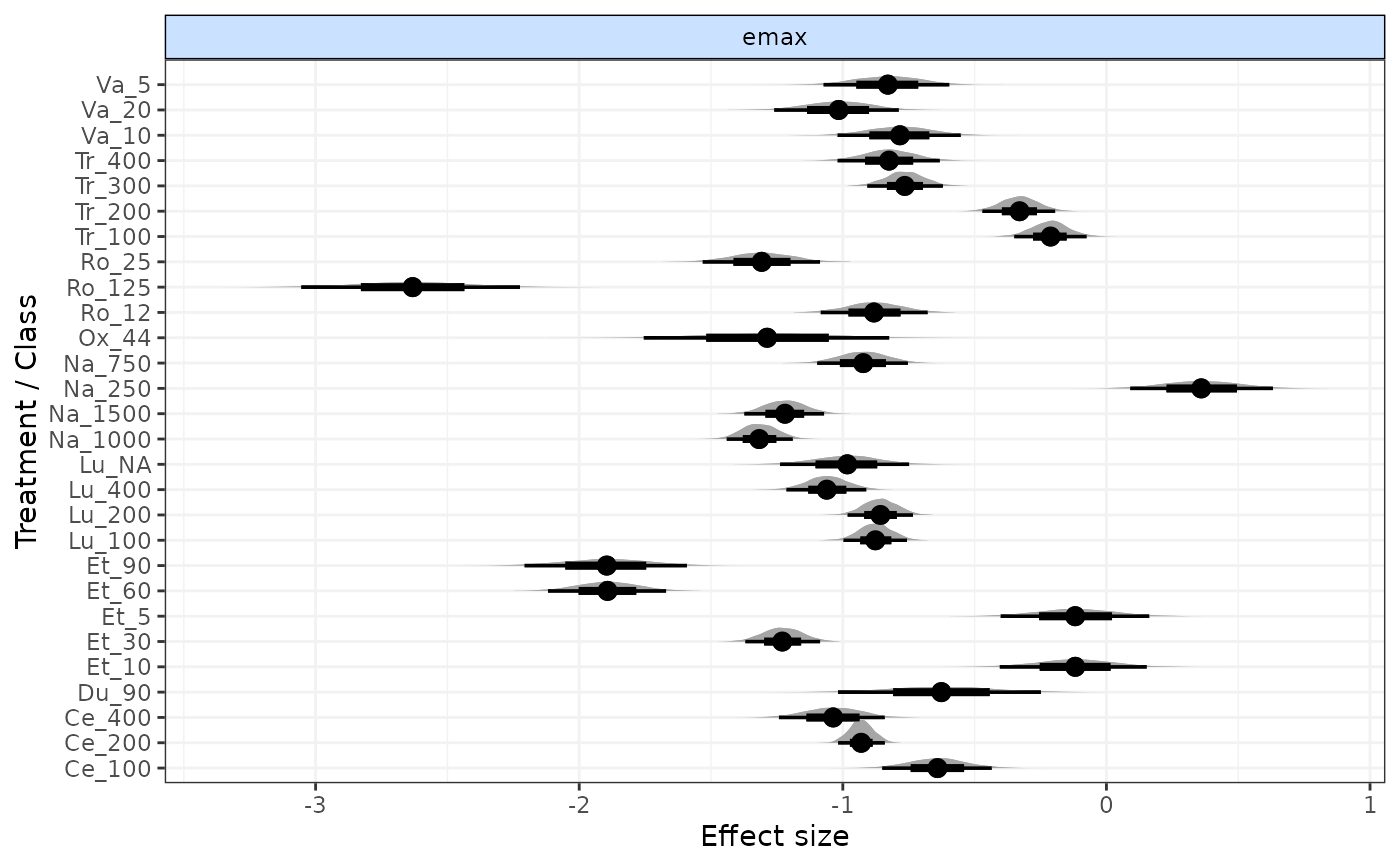
plot(result)
 ###### Additional model arguments ######
# Use gout dataset
goutnet <- mb.network(goutSUA_CFBcomb)
#> Reference treatment is `Plac`
#> Studies reporting change from baseline automatically identified from the data
# Define a user-defined time-course relationship for use in mb.run
timecourse <- ~ exp(beta.1 * time) + (time^beta.2)
# Run model with:
# user-defined time-course function
# random relative effects on beta.1
# default common effects on beta.2
# default relative pooling on beta.1 and beta.2
# common class effect on beta.2
mb.run(goutnet, fun=tuser(fun=timecourse, method.1="random"),
class.effect=list(beta.1="common"))
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 224
#> Unobserved stochastic nodes: 121
#> Total graph size: 4851
#>
#> Initializing model
#>
#> Inference for Bugs model at "/tmp/RtmpPWTcs6/file1eda187d45f8", fit using jags,
#> 3 chains, each with 20000 iterations (first 10000 discarded), n.thin = 10
#> n.sims = 3000 iterations saved. Running time = 34.452 secs
#> mu.vect sd.vect 2.5% 25% 50% 75%
#> D.1[1] 0.000 0.000 0.000 0.000 0.000 0.000
#> D.1[2] -2.118 20.473 -45.968 -14.182 -3.444 10.255
#> D.1[3] -16.930 24.838 -67.808 -31.971 -15.310 -5.130
#> D.1[4] 2.646 28.421 -42.129 -19.079 0.001 20.356
#> D.1[5] -11.735 24.160 -59.798 -27.102 -10.668 4.538
#> D.1[6] -22.912 12.563 -53.503 -29.500 -21.499 -14.146
#> D.1[7] -25.003 18.966 -67.405 -36.798 -25.581 -13.029
#> d.1[1] 0.000 0.000 0.000 0.000 0.000 0.000
#> d.1[2] -2.118 20.473 -45.968 -14.182 -3.444 10.255
#> d.1[3] -2.118 20.473 -45.968 -14.182 -3.444 10.255
#> d.1[4] -2.118 20.473 -45.968 -14.182 -3.444 10.255
#> d.1[5] -2.118 20.473 -45.968 -14.182 -3.444 10.255
#> d.1[6] -16.930 24.838 -67.808 -31.971 -15.310 -5.130
#> d.1[7] 2.646 28.421 -42.129 -19.079 0.001 20.356
#> d.1[8] 2.646 28.421 -42.129 -19.079 0.001 20.356
#> d.1[9] 2.646 28.421 -42.129 -19.079 0.001 20.356
#> d.1[10] 2.646 28.421 -42.129 -19.079 0.001 20.356
#> d.1[11] -11.735 24.160 -59.798 -27.102 -10.668 4.538
#> d.1[12] -22.912 12.563 -53.503 -29.500 -21.499 -14.146
#> d.1[13] -22.912 12.563 -53.503 -29.500 -21.499 -14.146
#> d.1[14] -22.912 12.563 -53.503 -29.500 -21.499 -14.146
#> d.1[15] -22.912 12.563 -53.503 -29.500 -21.499 -14.146
#> d.1[16] -25.003 18.966 -67.405 -36.798 -25.581 -13.029
#> d.1[17] -25.003 18.966 -67.405 -36.798 -25.581 -13.029
#> d.1[18] -25.003 18.966 -67.405 -36.798 -25.581 -13.029
#> d.1[19] -25.003 18.966 -67.405 -36.798 -25.581 -13.029
#> d.2[1] 0.000 0.000 0.000 0.000 0.000 0.000
#> d.2[2] -4.258 30.121 -64.362 -23.988 -3.908 15.377
#> d.2[3] 2.787 29.991 -55.663 -17.703 2.043 23.222
#> d.2[4] -16.216 10.037 -42.182 -21.185 -13.726 -8.763
#> d.2[5] 3.251 30.239 -55.131 -16.629 2.790 23.528
#> d.2[6] -15.286 28.572 -68.523 -33.332 -17.036 0.666
#> d.2[7] -3.130 29.594 -61.992 -23.073 -2.284 16.988
#> d.2[8] -8.295 27.617 -64.693 -26.056 -7.525 9.398
#> d.2[9] -2.159 30.253 -62.801 -22.088 -2.312 17.542
#> d.2[10] -5.797 27.870 -60.764 -24.387 -5.129 13.541
#> d.2[11] -16.581 25.050 -68.613 -32.505 -16.125 -0.400
#> d.2[12] -11.477 27.519 -68.633 -28.271 -10.667 6.432
#> d.2[13] -7.374 28.075 -64.858 -25.710 -6.914 11.592
#> d.2[14] -15.880 22.321 -64.696 -29.541 -14.189 -1.010
#> d.2[15] -16.294 24.557 -67.466 -31.895 -15.595 0.514
#> d.2[16] 4.312 30.092 -54.318 -15.397 3.837 23.443
#> d.2[17] -16.430 25.935 -68.444 -31.864 -16.255 -1.843
#> d.2[18] -15.111 23.712 -63.893 -29.706 -15.057 -0.880
#> d.2[19] -16.574 26.640 -69.535 -33.124 -16.911 -0.236
#> rho 0.000 0.000 0.000 0.000 0.000 0.000
#> sd.beta.1 3.162 2.226 0.167 1.377 2.760 4.569
#> totresdev 2013067.918 7.929 2013056.566 2013062.633 2013065.271 2013071.465
#> deviance 2012373.729 7.929 2012362.377 2012368.445 2012371.082 2012377.277
#> 97.5% Rhat n.eff
#> D.1[1] 0.000 1.000 1
#> D.1[2] 37.993 1.094 26
#> D.1[3] 40.861 1.327 11
#> D.1[4] 67.607 1.121 39
#> D.1[5] 35.313 1.056 44
#> D.1[6] -3.048 1.094 51
#> D.1[7] 12.685 1.054 49
#> d.1[1] 0.000 1.000 1
#> d.1[2] 37.993 1.094 26
#> d.1[3] 37.993 1.094 26
#> d.1[4] 37.993 1.094 26
#> d.1[5] 37.993 1.094 26
#> d.1[6] 40.861 1.327 11
#> d.1[7] 67.607 1.121 39
#> d.1[8] 67.607 1.121 39
#> d.1[9] 67.607 1.121 39
#> d.1[10] 67.607 1.121 39
#> d.1[11] 35.313 1.056 44
#> d.1[12] -3.048 1.094 51
#> d.1[13] -3.048 1.094 51
#> d.1[14] -3.048 1.094 51
#> d.1[15] -3.048 1.094 51
#> d.1[16] 12.685 1.054 49
#> d.1[17] 12.685 1.054 49
#> d.1[18] 12.685 1.054 49
#> d.1[19] 12.685 1.054 49
#> d.2[1] 0.000 1.000 1
#> d.2[2] 53.906 1.002 1900
#> d.2[3] 60.605 1.001 3000
#> d.2[4] -4.412 1.001 2100
#> d.2[5] 63.861 1.001 3000
#> d.2[6] 46.050 1.122 22
#> d.2[7] 52.753 1.001 3000
#> d.2[8] 44.331 1.002 1600
#> d.2[9] 58.187 1.001 3000
#> d.2[10] 47.087 1.002 1600
#> d.2[11] 31.086 1.003 690
#> d.2[12] 40.577 1.001 3000
#> d.2[13] 45.125 1.006 370
#> d.2[14] 25.469 1.001 3000
#> d.2[15] 29.584 1.001 2300
#> d.2[16] 65.261 1.001 3000
#> d.2[17] 37.224 1.126 22
#> d.2[18] 32.415 1.138 20
#> d.2[19] 37.934 1.162 17
#> rho 0.000 1.000 1
#> sd.beta.1 8.259 1.060 73
#> totresdev 2013085.220 1.000 1
#> deviance 2012391.032 1.000 1
#>
#> For each parameter, n.eff is a crude measure of effective sample size,
#> and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
#>
#> DIC info (using the rule: pV = var(deviance)/2)
#> pV = 28.8 and DIC = 2012379.7
#> DIC info (using the rule: pD = Dbar-Dhat, computed via 'rjags::dic.samples')
#> pD = 8.6 and DIC = 2012382.9
#> DIC is an estimate of expected predictive error (lower deviance is better).
# Fit a log-linear MBNMA
# with variance adjustment for correlation between time-points
result <- mb.run(network, fun=tloglin(),
rho="dunif(0,1)", covar="varadj")
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 417
#> Unobserved stochastic nodes: 89
#> Total graph size: 7303
#>
#> Initializing model
#>
# }
###### Additional model arguments ######
# Use gout dataset
goutnet <- mb.network(goutSUA_CFBcomb)
#> Reference treatment is `Plac`
#> Studies reporting change from baseline automatically identified from the data
# Define a user-defined time-course relationship for use in mb.run
timecourse <- ~ exp(beta.1 * time) + (time^beta.2)
# Run model with:
# user-defined time-course function
# random relative effects on beta.1
# default common effects on beta.2
# default relative pooling on beta.1 and beta.2
# common class effect on beta.2
mb.run(goutnet, fun=tuser(fun=timecourse, method.1="random"),
class.effect=list(beta.1="common"))
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 224
#> Unobserved stochastic nodes: 121
#> Total graph size: 4851
#>
#> Initializing model
#>
#> Inference for Bugs model at "/tmp/RtmpPWTcs6/file1eda187d45f8", fit using jags,
#> 3 chains, each with 20000 iterations (first 10000 discarded), n.thin = 10
#> n.sims = 3000 iterations saved. Running time = 34.452 secs
#> mu.vect sd.vect 2.5% 25% 50% 75%
#> D.1[1] 0.000 0.000 0.000 0.000 0.000 0.000
#> D.1[2] -2.118 20.473 -45.968 -14.182 -3.444 10.255
#> D.1[3] -16.930 24.838 -67.808 -31.971 -15.310 -5.130
#> D.1[4] 2.646 28.421 -42.129 -19.079 0.001 20.356
#> D.1[5] -11.735 24.160 -59.798 -27.102 -10.668 4.538
#> D.1[6] -22.912 12.563 -53.503 -29.500 -21.499 -14.146
#> D.1[7] -25.003 18.966 -67.405 -36.798 -25.581 -13.029
#> d.1[1] 0.000 0.000 0.000 0.000 0.000 0.000
#> d.1[2] -2.118 20.473 -45.968 -14.182 -3.444 10.255
#> d.1[3] -2.118 20.473 -45.968 -14.182 -3.444 10.255
#> d.1[4] -2.118 20.473 -45.968 -14.182 -3.444 10.255
#> d.1[5] -2.118 20.473 -45.968 -14.182 -3.444 10.255
#> d.1[6] -16.930 24.838 -67.808 -31.971 -15.310 -5.130
#> d.1[7] 2.646 28.421 -42.129 -19.079 0.001 20.356
#> d.1[8] 2.646 28.421 -42.129 -19.079 0.001 20.356
#> d.1[9] 2.646 28.421 -42.129 -19.079 0.001 20.356
#> d.1[10] 2.646 28.421 -42.129 -19.079 0.001 20.356
#> d.1[11] -11.735 24.160 -59.798 -27.102 -10.668 4.538
#> d.1[12] -22.912 12.563 -53.503 -29.500 -21.499 -14.146
#> d.1[13] -22.912 12.563 -53.503 -29.500 -21.499 -14.146
#> d.1[14] -22.912 12.563 -53.503 -29.500 -21.499 -14.146
#> d.1[15] -22.912 12.563 -53.503 -29.500 -21.499 -14.146
#> d.1[16] -25.003 18.966 -67.405 -36.798 -25.581 -13.029
#> d.1[17] -25.003 18.966 -67.405 -36.798 -25.581 -13.029
#> d.1[18] -25.003 18.966 -67.405 -36.798 -25.581 -13.029
#> d.1[19] -25.003 18.966 -67.405 -36.798 -25.581 -13.029
#> d.2[1] 0.000 0.000 0.000 0.000 0.000 0.000
#> d.2[2] -4.258 30.121 -64.362 -23.988 -3.908 15.377
#> d.2[3] 2.787 29.991 -55.663 -17.703 2.043 23.222
#> d.2[4] -16.216 10.037 -42.182 -21.185 -13.726 -8.763
#> d.2[5] 3.251 30.239 -55.131 -16.629 2.790 23.528
#> d.2[6] -15.286 28.572 -68.523 -33.332 -17.036 0.666
#> d.2[7] -3.130 29.594 -61.992 -23.073 -2.284 16.988
#> d.2[8] -8.295 27.617 -64.693 -26.056 -7.525 9.398
#> d.2[9] -2.159 30.253 -62.801 -22.088 -2.312 17.542
#> d.2[10] -5.797 27.870 -60.764 -24.387 -5.129 13.541
#> d.2[11] -16.581 25.050 -68.613 -32.505 -16.125 -0.400
#> d.2[12] -11.477 27.519 -68.633 -28.271 -10.667 6.432
#> d.2[13] -7.374 28.075 -64.858 -25.710 -6.914 11.592
#> d.2[14] -15.880 22.321 -64.696 -29.541 -14.189 -1.010
#> d.2[15] -16.294 24.557 -67.466 -31.895 -15.595 0.514
#> d.2[16] 4.312 30.092 -54.318 -15.397 3.837 23.443
#> d.2[17] -16.430 25.935 -68.444 -31.864 -16.255 -1.843
#> d.2[18] -15.111 23.712 -63.893 -29.706 -15.057 -0.880
#> d.2[19] -16.574 26.640 -69.535 -33.124 -16.911 -0.236
#> rho 0.000 0.000 0.000 0.000 0.000 0.000
#> sd.beta.1 3.162 2.226 0.167 1.377 2.760 4.569
#> totresdev 2013067.918 7.929 2013056.566 2013062.633 2013065.271 2013071.465
#> deviance 2012373.729 7.929 2012362.377 2012368.445 2012371.082 2012377.277
#> 97.5% Rhat n.eff
#> D.1[1] 0.000 1.000 1
#> D.1[2] 37.993 1.094 26
#> D.1[3] 40.861 1.327 11
#> D.1[4] 67.607 1.121 39
#> D.1[5] 35.313 1.056 44
#> D.1[6] -3.048 1.094 51
#> D.1[7] 12.685 1.054 49
#> d.1[1] 0.000 1.000 1
#> d.1[2] 37.993 1.094 26
#> d.1[3] 37.993 1.094 26
#> d.1[4] 37.993 1.094 26
#> d.1[5] 37.993 1.094 26
#> d.1[6] 40.861 1.327 11
#> d.1[7] 67.607 1.121 39
#> d.1[8] 67.607 1.121 39
#> d.1[9] 67.607 1.121 39
#> d.1[10] 67.607 1.121 39
#> d.1[11] 35.313 1.056 44
#> d.1[12] -3.048 1.094 51
#> d.1[13] -3.048 1.094 51
#> d.1[14] -3.048 1.094 51
#> d.1[15] -3.048 1.094 51
#> d.1[16] 12.685 1.054 49
#> d.1[17] 12.685 1.054 49
#> d.1[18] 12.685 1.054 49
#> d.1[19] 12.685 1.054 49
#> d.2[1] 0.000 1.000 1
#> d.2[2] 53.906 1.002 1900
#> d.2[3] 60.605 1.001 3000
#> d.2[4] -4.412 1.001 2100
#> d.2[5] 63.861 1.001 3000
#> d.2[6] 46.050 1.122 22
#> d.2[7] 52.753 1.001 3000
#> d.2[8] 44.331 1.002 1600
#> d.2[9] 58.187 1.001 3000
#> d.2[10] 47.087 1.002 1600
#> d.2[11] 31.086 1.003 690
#> d.2[12] 40.577 1.001 3000
#> d.2[13] 45.125 1.006 370
#> d.2[14] 25.469 1.001 3000
#> d.2[15] 29.584 1.001 2300
#> d.2[16] 65.261 1.001 3000
#> d.2[17] 37.224 1.126 22
#> d.2[18] 32.415 1.138 20
#> d.2[19] 37.934 1.162 17
#> rho 0.000 1.000 1
#> sd.beta.1 8.259 1.060 73
#> totresdev 2013085.220 1.000 1
#> deviance 2012391.032 1.000 1
#>
#> For each parameter, n.eff is a crude measure of effective sample size,
#> and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
#>
#> DIC info (using the rule: pV = var(deviance)/2)
#> pV = 28.8 and DIC = 2012379.7
#> DIC info (using the rule: pD = Dbar-Dhat, computed via 'rjags::dic.samples')
#> pD = 8.6 and DIC = 2012382.9
#> DIC is an estimate of expected predictive error (lower deviance is better).
# Fit a log-linear MBNMA
# with variance adjustment for correlation between time-points
result <- mb.run(network, fun=tloglin(),
rho="dunif(0,1)", covar="varadj")
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 417
#> Unobserved stochastic nodes: 89
#> Total graph size: 7303
#>
#> Initializing model
#>
# }