Rank relative effects obtained between specific doses
rank.relative.array.RdRanks "relative.table" objects generated by get.relative().
# S3 method for class 'relative.array'
rank(x, lower_better = TRUE, ...)Arguments
Value
An object of class("mbnma.rank") which is a list containing a summary data
frame, a matrix of rankings for each MCMC iteration, and a matrix of probabilities
that each agent has a particular rank, for each parameter that has been ranked.
Examples
# \donttest{
# Using the triptans data
network <- mbnma.network(triptans)
#> Values for `agent` with dose = 0 have been recoded to `Placebo`
#> agent is being recoded to enforce sequential numbering
# Rank selected predictions from an Emax dose-response MBNMA
emax <- mbnma.run(network, fun=demax(), method="random")
#> `likelihood` not given by user - set to `binomial` based on data provided
#> `link` not given by user - set to `logit` based on assigned value for `likelihood`
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 182
#> Unobserved stochastic nodes: 197
#> Total graph size: 4115
#>
#> Initializing model
#>
rels <- get.relative(emax)
rank <- rank(rels, lower_better=TRUE)
# Print and generate summary data frame for `mbnma.rank` object
summary(rank)
#> $RelativeEffects
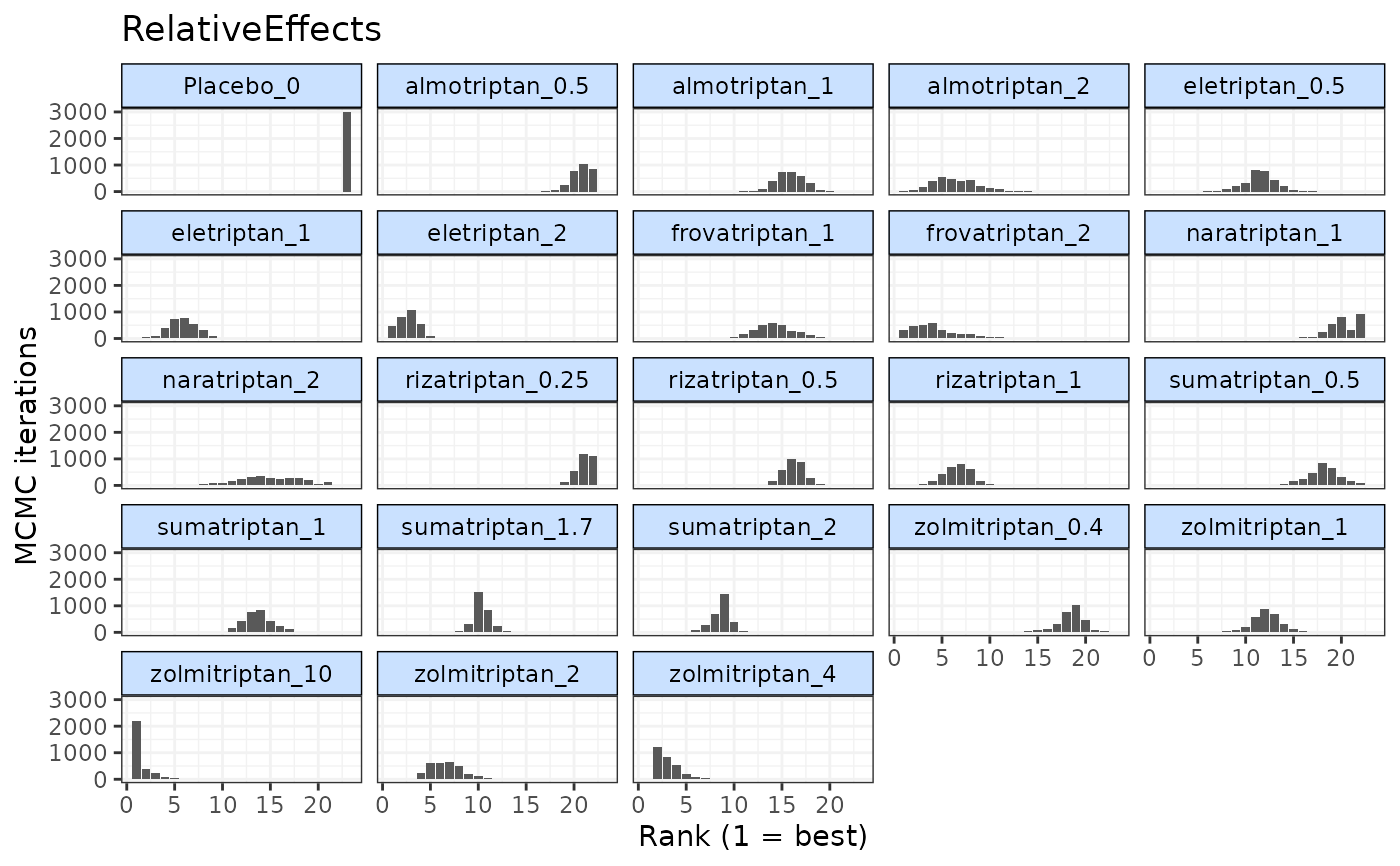
#> rank.param mean sd 2.5% 25% 50% 75% 97.5%
#> 1 Placebo_0 23.000000 0.0000000 23 23 23 23 23
#> 2 eletriptan_0.5 11.497667 1.6340802 8 11 12 12 15
#> 3 eletriptan_1 5.784667 1.4670886 3 5 6 7 9
#> 4 eletriptan_2 2.581333 1.0766172 1 2 3 3 5
#> 5 sumatriptan_0.5 17.953667 1.5518771 15 17 18 19 21
#> 6 sumatriptan_1 13.584000 1.3312405 11 13 14 14 16
#> 7 sumatriptan_1.7 10.273667 0.9134466 8 10 10 11 12
#> 8 sumatriptan_2 8.657667 1.0441127 6 8 9 9 10
#> 9 frovatriptan_1 14.123667 2.1342973 10 13 14 15 19
#> 10 frovatriptan_2 4.197667 2.6116792 1 2 4 5 11
#> 11 almotriptan_0.5 20.826333 0.9628857 19 20 21 22 22
#> 12 almotriptan_1 15.900000 1.4470768 13 15 16 17 19
#> 13 almotriptan_2 6.220000 2.2201899 2 5 6 8 11
#> 14 zolmitriptan_0.4 18.322000 1.4478771 15 18 19 19 21
#> 15 zolmitriptan_1 12.127667 1.4177581 9 11 12 13 15
#> 16 zolmitriptan_2 6.658000 1.6554807 4 5 7 8 10
#> 17 zolmitriptan_4 3.318000 1.4556496 2 2 3 4 7
#> 18 zolmitriptan_10 1.621000 1.1896067 1 1 1 2 5
#> 19 naratriptan_1 20.319333 1.4805709 17 19 20 22 22
#> 20 naratriptan_2 14.835333 3.3933351 8 13 15 17 21
#> 21 rizatriptan_0.25 21.163000 0.8070407 19 21 21 22 22
#> 22 rizatriptan_0.5 16.343333 1.0987226 14 16 16 17 18
#> 23 rizatriptan_1 6.692000 1.4759840 4 6 7 8 9
#>
print(rank)
#>
#> ================================
#> Ranking of dose-response MBNMA
#> ================================
#>
#> Includes ranking of relative effects
#>
#> 23 relefs ranked with negative responses being `worse`
#>
#> RelativeEffects ranking (from best to worst)
#>
#> |Treatment | Mean| Median| 2.5%| 97.5%|
#> |:----------------|-----:|------:|----:|-----:|
#> |zolmitriptan_10 | 1.62| 1| 1| 5|
#> |eletriptan_2 | 2.58| 3| 1| 5|
#> |zolmitriptan_4 | 3.32| 3| 2| 7|
#> |frovatriptan_2 | 4.20| 4| 1| 11|
#> |eletriptan_1 | 5.78| 6| 3| 9|
#> |almotriptan_2 | 6.22| 6| 2| 11|
#> |zolmitriptan_2 | 6.66| 7| 4| 10|
#> |rizatriptan_1 | 6.69| 7| 4| 9|
#> |sumatriptan_2 | 8.66| 9| 6| 10|
#> |sumatriptan_1.7 | 10.27| 10| 8| 12|
#> |eletriptan_0.5 | 11.50| 12| 8| 15|
#> |zolmitriptan_1 | 12.13| 12| 9| 15|
#> |sumatriptan_1 | 13.58| 14| 11| 16|
#> |frovatriptan_1 | 14.12| 14| 10| 19|
#> |naratriptan_2 | 14.84| 15| 8| 21|
#> |almotriptan_1 | 15.90| 16| 13| 19|
#> |rizatriptan_0.5 | 16.34| 16| 14| 18|
#> |sumatriptan_0.5 | 17.95| 18| 15| 21|
#> |zolmitriptan_0.4 | 18.32| 19| 15| 21|
#> |naratriptan_1 | 20.32| 20| 17| 22|
#> |almotriptan_0.5 | 20.83| 21| 19| 22|
#> |rizatriptan_0.25 | 21.16| 21| 19| 22|
#> |Placebo_0 | 23.00| 23| 23| 23|
#>
#>
# Plot `mbnma.rank` object
plot(rank)
 # }
# }