MBNMAdose: Perform Network Meta-Regression
Hugo Pedder
2025-05-22
metaregression-6.RmdNetwork meta-regression
Study-level covariates can be included in the model to adjust treatment effects following an approach for meta-regression outlined in NICE Technical Support Document 3 (Dias et al. 2011). This can be used to explore and account for potential effect modification.
Following the definition in NICE Technical Support Document 3, network meta-regression can be expressed as an interaction on the treatment effect in arms :
where is the linear predictor, is the baseline effect on arm 1, is the dose-response function at dose with dose-response parameters for agent in arm of study . is then the effect modifying interaction between the agent in arm and the network reference agent (typically Placebo in a dose-response analysis).
Data preparation
To improve estimation:
- continuous covariates should be centred around their mean
- binary/categorical variables should be recoded with the most commonly reported value as the reference category
# Using the SSRI dataset
ssri.reg <- ssri
# For a continuous covariate
ssri.reg <- ssri.reg %>%
dplyr::mutate(x.weeks = weeks - mean(weeks, na.rm = TRUE))
# For a categorical covariate
table(ssri$weeks) # Using 8 weeks as the reference
ssri.reg <- ssri.reg %>%
dplyr::mutate(r.weeks = factor(weeks, levels = c(8, 4, 5, 6, 9, 10)))
# Create network object
ssrinet <- mbnma.network(ssri.reg)
#> Values for `agent` with dose = 0 have been recoded to `Placebo`
#> agent is being recoded to enforce sequential numberingModelling
For performing network meta-regression, different assumptions can be made regarding how the effect modification may be shared across agents:
Independent, agent-specific interactions
The least constraining assumption available in MBNMAdose
is to assume that the effect modifier acts on each agent independently,
and separate
are therefore estimated for each agent in the network.
A slightly stronger assumption is to assume that agents within the same class share the same interaction effect, though classes must be specified within the dataset for this.
# Regress for continuous weeks Separate effect modification for each agent vs
# Placebo
ssrimod.a <- mbnma.run(ssrinet, fun = dfpoly(degree = 2), regress = ~x.weeks, regress.effect = "agent")
summary(ssrimod.a)
#> ========================================
#> Dose-response MBNMA
#> ========================================
#>
#> Likelihood: binomial
#> Link function: logit
#> Dose-response function: fpoly
#>
#> Pooling method
#>
#> Method: Common (fixed) effects estimated for relative effects
#>
#>
#> beta.1 dose-response parameter results
#>
#> Pooling: relative effects for each agent
#>
#> |Agent |Parameter | Median| 2.5%| 97.5%|
#> |:------------|:---------|-------:|-------:|-------:|
#> |citalopram |beta.1[2] | -0.0905| -0.3472| 0.1787|
#> |escitalopram |beta.1[3] | 0.1313| -0.1387| 0.4076|
#> |fluoxetine |beta.1[4] | 0.2670| 0.0301| 0.4954|
#> |paroxetine |beta.1[5] | -0.1325| -0.4393| 0.1681|
#> |sertraline |beta.1[6] | 1.2071| -8.0971| 10.5708|
#>
#>
#> beta.2 dose-response parameter results
#>
#> Pooling: relative effects for each agent
#>
#> |Agent |Parameter | Median| 2.5%| 97.5%|
#> |:------------|:---------|-------:|-------:|------:|
#> |citalopram |beta.2[2] | 0.0664| -0.0047| 0.1384|
#> |escitalopram |beta.2[3] | -0.0062| -0.0943| 0.0782|
#> |fluoxetine |beta.2[4] | -0.0653| -0.1316| 0.0031|
#> |paroxetine |beta.2[5] | 0.0898| -0.0050| 0.1884|
#> |sertraline |beta.2[6] | -0.1688| -1.1868| 0.8442|
#>
#>
#> power.1 dose-response parameter results
#>
#> Assigned a numeric value: 0
#>
#> power.2 dose-response parameter results
#>
#> Assigned a numeric value: 0
#>
#> Meta-regression
#>
#> Covariates interacting with study-level relative effects: x.weeks
#> Common (identical) covariate-by-agent effects
#>
#>
#> |Agent |Parameter | Median| 2.5%| 97.5%|
#> |:------------|:------------|-------:|--------:|-------:|
#> |citalopram |B.x.weeks[2] | -0.1601| -0.3384| 0.0164|
#> |escitalopram |B.x.weeks[3] | 0.1603| -0.0356| 0.3581|
#> |fluoxetine |B.x.weeks[4] | -0.0316| -0.1713| 0.1084|
#> |paroxetine |B.x.weeks[5] | 0.0862| -0.0235| 0.1936|
#> |sertraline |B.x.weeks[6] | 1.2913| -18.3645| 20.9489|
#>
#>
#> Model Fit Statistics
#> Deviance = 890.8
#> Residual deviance = 191.1
#> Deviance Information Criterion (DIC) = 966.4Within the output, a separate parameter (named
B.x.weeks[]) has been estimated for each agent that
corresponds to the effect of an additional week of study follow-up on
the relative effect of the agent versus Placebo. Note that due to the
inclusion of weeks as a continuous covariate, we are assuming a linear
effect modification due to study follow-up.
Random effect interaction
Alternatively, the effect modification for different agents versus the network reference agent can be assumed to be exchangeable/shared across the network about a common mean, , with a between-agent standard deviation of :
# Regress for continuous weeks Random effect modification across all agents vs
# Placebo
ssrimod.r <- mbnma.run(ssrinet, fun = dfpoly(degree = 2), regress = ~x.weeks, regress.effect = "random")
summary(ssrimod.r)
#> ========================================
#> Dose-response MBNMA
#> ========================================
#>
#> Likelihood: binomial
#> Link function: logit
#> Dose-response function: fpoly
#>
#> Pooling method
#>
#> Method: Common (fixed) effects estimated for relative effects
#>
#>
#> beta.1 dose-response parameter results
#>
#> Pooling: relative effects for each agent
#>
#> |Agent |Parameter | Median| 2.5%| 97.5%|
#> |:------------|:---------|-------:|-------:|------:|
#> |citalopram |beta.1[2] | -0.0508| -0.4208| 0.2349|
#> |escitalopram |beta.1[3] | 0.2452| -0.0534| 0.5715|
#> |fluoxetine |beta.1[4] | 0.2807| 0.0184| 0.5190|
#> |paroxetine |beta.1[5] | -0.1117| -0.4571| 0.2104|
#> |sertraline |beta.1[6] | 0.6415| 0.1647| 1.2119|
#>
#>
#> beta.2 dose-response parameter results
#>
#> Pooling: relative effects for each agent
#>
#> |Agent |Parameter | Median| 2.5%| 97.5%|
#> |:------------|:---------|-------:|-------:|-------:|
#> |citalopram |beta.2[2] | 0.0572| -0.0212| 0.1604|
#> |escitalopram |beta.2[3] | -0.0358| -0.1457| 0.0652|
#> |fluoxetine |beta.2[4] | -0.0681| -0.1375| 0.0123|
#> |paroxetine |beta.2[5] | 0.0876| -0.0148| 0.1970|
#> |sertraline |beta.2[6] | -0.1088| -0.2280| -0.0111|
#>
#>
#> power.1 dose-response parameter results
#>
#> Assigned a numeric value: 0
#>
#> power.2 dose-response parameter results
#>
#> Assigned a numeric value: 0
#>
#> Meta-regression
#>
#> Covariates interacting with study-level relative effects: x.weeks
#> Random (exchangeable) covariate-by-treatment effects
#>
#>
#> |Regression effect |Parameter | Median| 2.5%| 97.5%|
#> |:-----------------|:---------|------:|-------:|------:|
#> |Random effect |B.x.weeks | 0.0254| -0.0664| 0.1221|
#>
#>
#> Standard deviation for random covariate-by-treatment effects
#>
#> |Parameter | Median| 2.5%| 97.5%|
#> |:------------|------:|------:|------:|
#> |sd.B.x.weeks | 0.0637| 0.0026| 0.2297|
#>
#>
#> Model Fit Statistics
#> Deviance = 891.5
#> Residual deviance = 191.8
#> Deviance Information Criterion (DIC) = 980.4In this case only a single regression paramter is estimated
(B.x.weeks), which corresponds to the mean effect of an
additional week of study follow-up on the relative effect of an active
agent versus Placebo. A parameter is also estimated for the
between-agent standard deviation, sd.B.x.weeks.
Common effect interaction
This is the strongest assumption for network meta-regression, and it implies that effect modification is common (equal) for all agents versus the network reference agent:
# Regress for categorical weeks Common effect modification across all agents vs
# Placebo
ssrimod.c <- mbnma.run(ssrinet, fun = dfpoly(degree = 2), regress = ~r.weeks, regress.effect = "common")
summary(ssrimod.c)
#> ========================================
#> Dose-response MBNMA
#> ========================================
#>
#> Likelihood: binomial
#> Link function: logit
#> Dose-response function: fpoly
#>
#> Pooling method
#>
#> Method: Common (fixed) effects estimated for relative effects
#>
#>
#> beta.1 dose-response parameter results
#>
#> Pooling: relative effects for each agent
#>
#> |Agent |Parameter | Median| 2.5%| 97.5%|
#> |:------------|:---------|-------:|-------:|------:|
#> |citalopram |beta.1[2] | -0.0686| -0.3486| 0.1968|
#> |escitalopram |beta.1[3] | 0.2473| 0.0215| 0.4695|
#> |fluoxetine |beta.1[4] | 0.2193| -0.0241| 0.4693|
#> |paroxetine |beta.1[5] | -0.1097| -0.4053| 0.1955|
#> |sertraline |beta.1[6] | 0.5893| 0.1109| 1.0744|
#>
#>
#> beta.2 dose-response parameter results
#>
#> Pooling: relative effects for each agent
#>
#> |Agent |Parameter | Median| 2.5%| 97.5%|
#> |:------------|:---------|-------:|-------:|-------:|
#> |citalopram |beta.2[2] | 0.0592| -0.0127| 0.1338|
#> |escitalopram |beta.2[3] | -0.0288| -0.1054| 0.0488|
#> |fluoxetine |beta.2[4] | -0.0561| -0.1259| 0.0128|
#> |paroxetine |beta.2[5] | 0.0855| -0.0100| 0.1809|
#> |sertraline |beta.2[6] | -0.1017| -0.1979| -0.0047|
#>
#>
#> power.1 dose-response parameter results
#>
#> Assigned a numeric value: 0
#>
#> power.2 dose-response parameter results
#>
#> Assigned a numeric value: 0
#>
#> Meta-regression
#>
#> Covariates interacting with study-level relative effects: r.weeks4, r.weeks5, r.weeks6, r.weeks9, r.weeks10
#> Common (identical) covariate-by-treatment effects
#>
#>
#> |Regression effect |Parameter | Median| 2.5%| 97.5%|
#> |:-----------------|:-----------|-------:|-------:|-------:|
#> |Common effect |B.r.weeks10 | 0.1504| -0.2455| 0.5443|
#> |Common effect |B.r.weeks4 | -0.7459| -1.4320| -0.0630|
#> |Common effect |B.r.weeks5 | 0.8323| -0.1496| 1.8133|
#> |Common effect |B.r.weeks6 | 0.0535| -0.1387| 0.2462|
#> |Common effect |B.r.weeks9 | 0.4415| -0.3891| 1.3221|
#>
#>
#> Model Fit Statistics
#> Deviance = 891.2
#> Residual deviance = 191.5
#> Deviance Information Criterion (DIC) = 964.2In this case we have performed the network meta-regression on study follow-up (weeks) as a categorical covariate. Therefore, although only a single parameter is estimated for each effect modifying term, there is a separate term for each category of week and a linear relationship for effect modification is no longer assumed.
Alternative assumptions
Although this is beyond the capability of MBNMAdose, one
could envision a more complex model in which the interaction effect also
varied by a dose-response relationship, rather than assuming an effect
by agent/class or across the whole network. This would in principle
contain fewer parameters than a fully independent interaction model (in
which a separate regression covariate is estimated for each treatment in
the dataset).
Aggregation bias
Note that adjusting for aggregated patient-level covariates
(e.g. mean age, % males, etc.) whilst using a non-identity link function
can introduce aggregation bias. This is a form of ecological bias that
biases treatment effects towards the null and is typically more severe
where treatment effects are strong and where the link function is highly
non-linear (Dias et al. 2011). This can be
resolved by performing a patient-level regression, but Individual
Participant Data are required for this and such an analysis is outside
the scope of MBNMAdose.
Prediction using effect modifying covariates
Models fitted with meta-regression can also be used to make
predictions for a specified set of covariate values. This includes when
estimating relative effects using get.relative(). An
additional argument regress.vals can be used to provide a
named vector of covariate values at which to make predictions.
# For a continuous covariate, make predictions at 5 weeks follow-up
pred <- predict(ssrimod.a, regress.vals = c(x.weeks = 5))
plot(pred)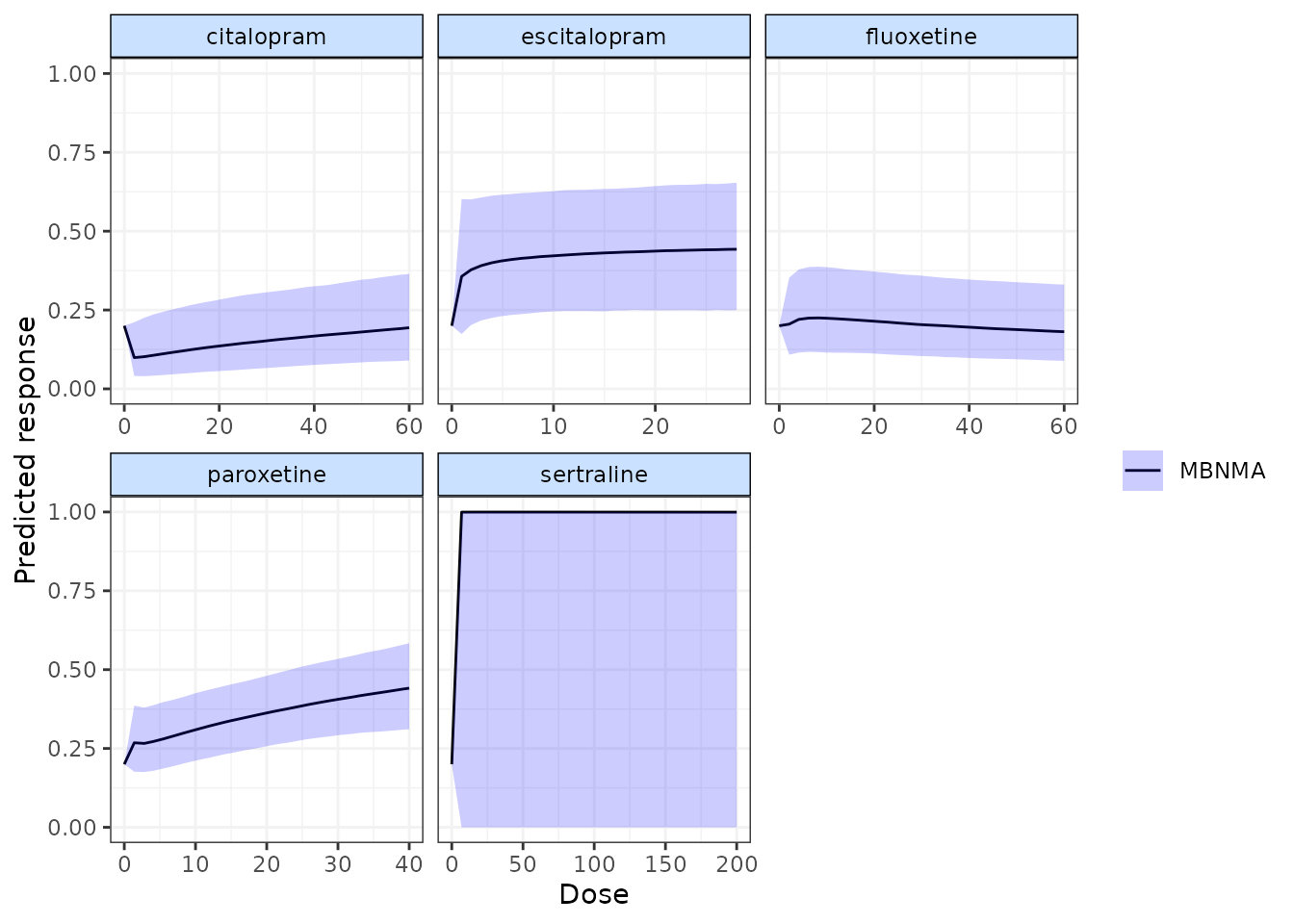
Predictions are very uncertain for Sertraline, as studies only investigated this agent at 6 weeks follow-up and therefore the agent-specific effect modification is very poorly estimated.
# For a categorical covariate, make predictions at 10 weeks follow-up
regress.p <- c(r.weeks10 = 1, r.weeks4 = 0, r.weeks5 = 0, r.weeks6 = 0, r.weeks9 = 0)
pred <- predict(ssrimod.c, regress.vals = regress.p)
plot(pred)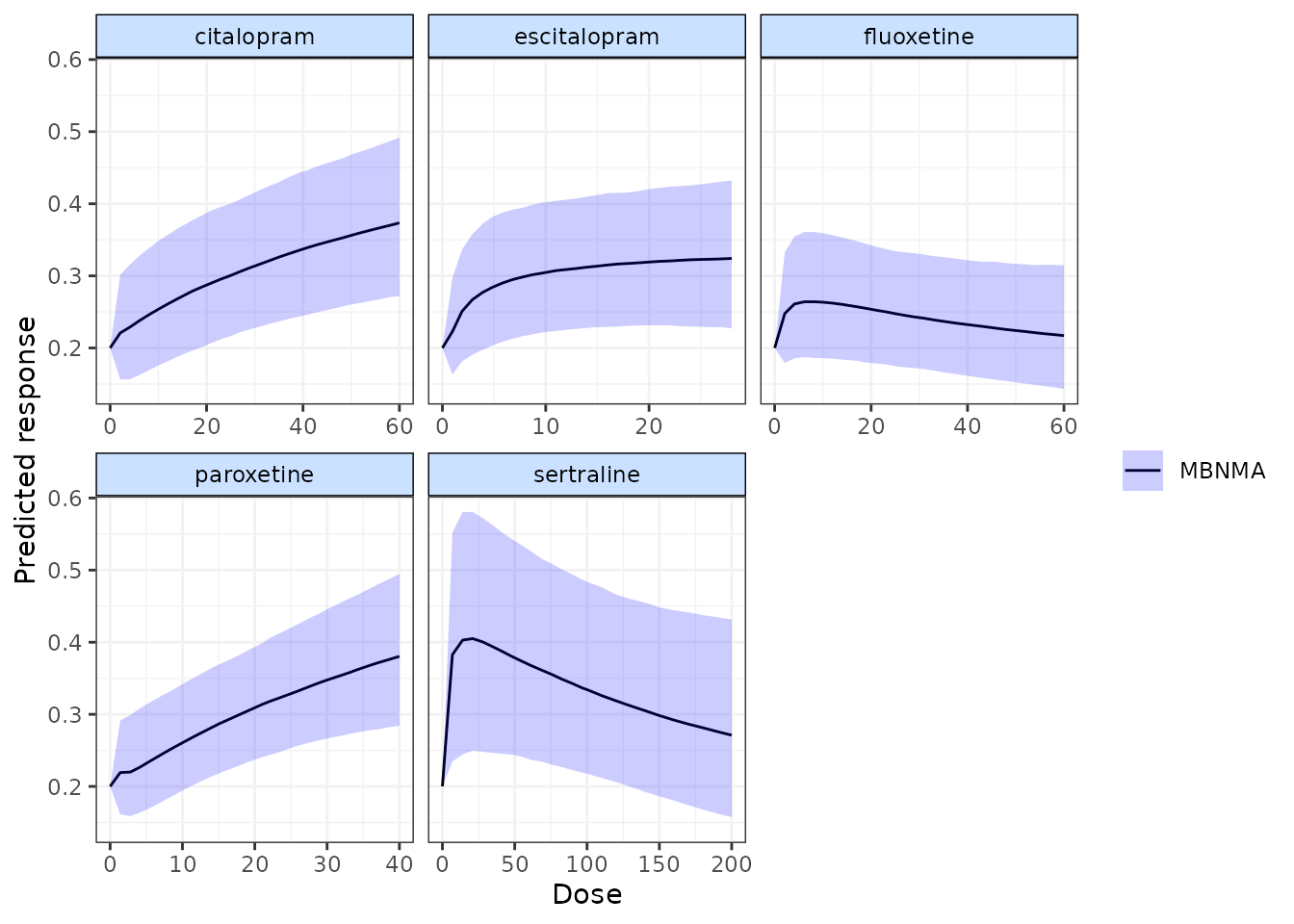
Note that categorical covariates are modelled as multiple binary dummy covariates, and so a value for each of these must be included.