Node-splitting model for testing consistency at the treatment-level
nma.nodesplit.RdSplits contributions for a given set of treatment comparisons into direct and indirect evidence. A discrepancy between the two suggests that the consistency assumption required for NMA (and subsequently MBNMA) may violated.
nma.nodesplit(
network,
likelihood = NULL,
link = NULL,
method = "common",
comparisons = NULL,
drop.discon = TRUE,
...
)
# S3 method for class 'nma.nodesplit'
plot(x, plot.type = NULL, ...)Arguments
- network
An object of class
mbnma.network.- likelihood
A string indicating the likelihood to use in the model. Can take either
"binomial","normal"or"poisson". If left asNULLthe likelihood will be inferred from the data.- link
A string indicating the link function to use in the model. Can take any link function defined within JAGS (e.g.
"logit","log","probit","cloglog"), be assigned the value"identity"for an identity link function, or be assigned the value"smd"for modelling Standardised Mean Differences using an identity link function. If left asNULLthe link function will be automatically assigned based on the likelihood.- method
Can take either
"common"or"random"to indicate whether relative effects should be modelled with between-study heterogeneity or not (see details).- comparisons
A matrix specifying the comparisons to be split (one row per comparison). The matrix must have two columns indicating each treatment for each comparison. Values can either be character (corresponding to the treatment names given in
network) or numeric (corresponding to treatment codes within thenetwork- note that these may change ifdrop.discon = TRUE).- drop.discon
A boolean object that indicates whether to drop treatments that are disconnected at the treatment level. Default is
TRUE. If set toFALSEthen this could lead to identification of nodesplit comparisons that are not connected to the network reference treatment, or lead to errors in running the nodesplit models, though it can be useful for error checking.- ...
Arguments to be sent to
ggplot2::ggplot()- x
An object of
class("nma.nodesplit")- plot.type
A character string that can take the value of
"forest"to plot only forest plots,"density"to plot only density plots, or left asNULL(the default) to plot both types of plot.
Value
Plots the desired graph(s) and returns an object (or list of object if
plot.type=NULL) of class(c("gg", "ggplot"))
Details
The S3 method plot() on an nma.nodesplit object generates either
forest plots of posterior medians and 95\% credible intervals, or density plots
of posterior densities for direct and indirect evidence.
Methods (by generic)
plot(nma.nodesplit): Plot outputs from treatment-level nodesplit models
Examples
# \donttest{
# Using the triptans data
network <- mbnma.network(triptans)
#> Values for `agent` with dose = 0 have been recoded to `Placebo`
#> agent is being recoded to enforce sequential numbering
split <- nma.nodesplit(network, likelihood = "binomial", link="logit",
method="common")
#>
|
| | 0%
|
|=== | 4%
|
|===== | 8%
|
|======== | 12%
|
|=========== | 15%
#>
|
|============= | 19%
#>
|
|================ | 23%
|
|=================== | 27%
|
|====================== | 31%
|
|======================== | 35%
|
|=========================== | 38%
|
|============================== | 42%
#>
|
|================================ | 46%
|
|=================================== | 50%
|
|====================================== | 54%
#>
|
|======================================== | 58%
|
|=========================================== | 62%
|
|============================================== | 65%
|
|================================================ | 69%
|
|=================================================== | 73%
|
|====================================================== | 77%
|
|========================================================= | 81%
#>
|
|=========================================================== | 85%
#>
|
|============================================================== | 88%
#>
|
|================================================================= | 92%
#>
|
|=================================================================== | 96%
#>
|
|======================================================================| 100%
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 182
#> Unobserved stochastic nodes: 92
#> Total graph size: 3436
#>
#> Initializing model
#>
#> [1] "Calculating nodesplit for: zolmitriptan_1 vs almotriptan_1"
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 180
#> Unobserved stochastic nodes: 91
#> Total graph size: 3397
#>
#> Initializing model
#>
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 182
#> Unobserved stochastic nodes: 323
#> Total graph size: 3641
#>
#> Initializing model
#>
#> [1] "Calculating nodesplit for: rizatriptan_1 vs sumatriptan_1"
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 180
#> Unobserved stochastic nodes: 91
#> Total graph size: 3395
#>
#> Initializing model
#>
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 182
#> Unobserved stochastic nodes: 323
#> Total graph size: 3641
#>
#> Initializing model
#>
#> [1] "Calculating nodesplit for: almotriptan_1 vs sumatriptan_1"
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 180
#> Unobserved stochastic nodes: 91
#> Total graph size: 3395
#>
#> Initializing model
#>
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 182
#> Unobserved stochastic nodes: 323
#> Total graph size: 3641
#>
#> Initializing model
#>
#> [1] "Calculating nodesplit for: rizatriptan_0.5 vs sumatriptan_0.5"
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 180
#> Unobserved stochastic nodes: 91
#> Total graph size: 3396
#>
#> Initializing model
#>
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 182
#> Unobserved stochastic nodes: 323
#> Total graph size: 3641
#>
#> Initializing model
#>
#### To perform nodesplit on selected comparisons ####
# Check for closed loops of treatments with independent evidence sources
loops <- inconsistency.loops(network$data.ab)
#>
|
| | 0%
|
|=== | 4%
|
|===== | 8%
|
|======== | 12%
|
|=========== | 15%
#>
|
|============= | 19%
#>
|
|================ | 23%
|
|=================== | 27%
|
|====================== | 31%
|
|======================== | 35%
|
|=========================== | 38%
|
|============================== | 42%
#>
|
|================================ | 46%
|
|=================================== | 50%
|
|====================================== | 54%
#>
|
|======================================== | 58%
|
|=========================================== | 62%
|
|============================================== | 65%
|
|================================================ | 69%
|
|=================================================== | 73%
|
|====================================================== | 77%
|
|========================================================= | 81%
#>
|
|=========================================================== | 85%
#>
|
|============================================================== | 88%
#>
|
|================================================================= | 92%
#>
|
|=================================================================== | 96%
#>
|
|======================================================================| 100%
# This...
single.split <- nma.nodesplit(network, likelihood = "binomial", link="logit",
method="random", comparisons=rbind(c("sumatriptan_1", "almotriptan_1")))
#>
|
| | 0%
|
|=== | 4%
|
|===== | 8%
|
|======== | 12%
|
|=========== | 15%
#>
|
|============= | 19%
#>
|
|================ | 23%
|
|=================== | 27%
|
|====================== | 31%
|
|======================== | 35%
|
|=========================== | 38%
|
|============================== | 42%
#>
|
|================================ | 46%
|
|=================================== | 50%
|
|====================================== | 54%
#>
|
|======================================== | 58%
|
|=========================================== | 62%
|
|============================================== | 65%
|
|================================================ | 69%
|
|=================================================== | 73%
|
|====================================================== | 77%
|
|========================================================= | 81%
#>
|
|=========================================================== | 85%
#>
|
|============================================================== | 88%
#>
|
|================================================================= | 92%
#>
|
|=================================================================== | 96%
#>
|
|======================================================================| 100%
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 182
#> Unobserved stochastic nodes: 205
#> Total graph size: 3979
#>
#> Initializing model
#>
#> [1] "Calculating nodesplit for: almotriptan_1 vs sumatriptan_1"
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 180
#> Unobserved stochastic nodes: 203
#> Total graph size: 3934
#>
#> Initializing model
#>
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 182
#> Unobserved stochastic nodes: 436
#> Total graph size: 4071
#>
#> Initializing model
#>
#...is the same as...
single.split <- nma.nodesplit(network, likelihood = "binomial", link="logit",
method="random", comparisons=rbind(c(6, 12)))
#>
|
| | 0%
|
|=== | 4%
|
|===== | 8%
|
|======== | 12%
|
|=========== | 15%
#>
|
|============= | 19%
#>
|
|================ | 23%
|
|=================== | 27%
|
|====================== | 31%
|
|======================== | 35%
|
|=========================== | 38%
|
|============================== | 42%
#>
|
|================================ | 46%
|
|=================================== | 50%
|
|====================================== | 54%
#>
|
|======================================== | 58%
|
|=========================================== | 62%
|
|============================================== | 65%
|
|================================================ | 69%
|
|=================================================== | 73%
|
|====================================================== | 77%
|
|========================================================= | 81%
#>
|
|=========================================================== | 85%
#>
|
|============================================================== | 88%
#>
|
|================================================================= | 92%
#>
|
|=================================================================== | 96%
#>
|
|======================================================================| 100%
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 182
#> Unobserved stochastic nodes: 205
#> Total graph size: 3979
#>
#> Initializing model
#>
#> [1] "Calculating nodesplit for: almotriptan_1 vs sumatriptan_1"
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 180
#> Unobserved stochastic nodes: 203
#> Total graph size: 3934
#>
#> Initializing model
#>
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 182
#> Unobserved stochastic nodes: 436
#> Total graph size: 4071
#>
#> Initializing model
#>
# Plot results
plot(split, plot.type="density") # Plot density plots of posterior densities
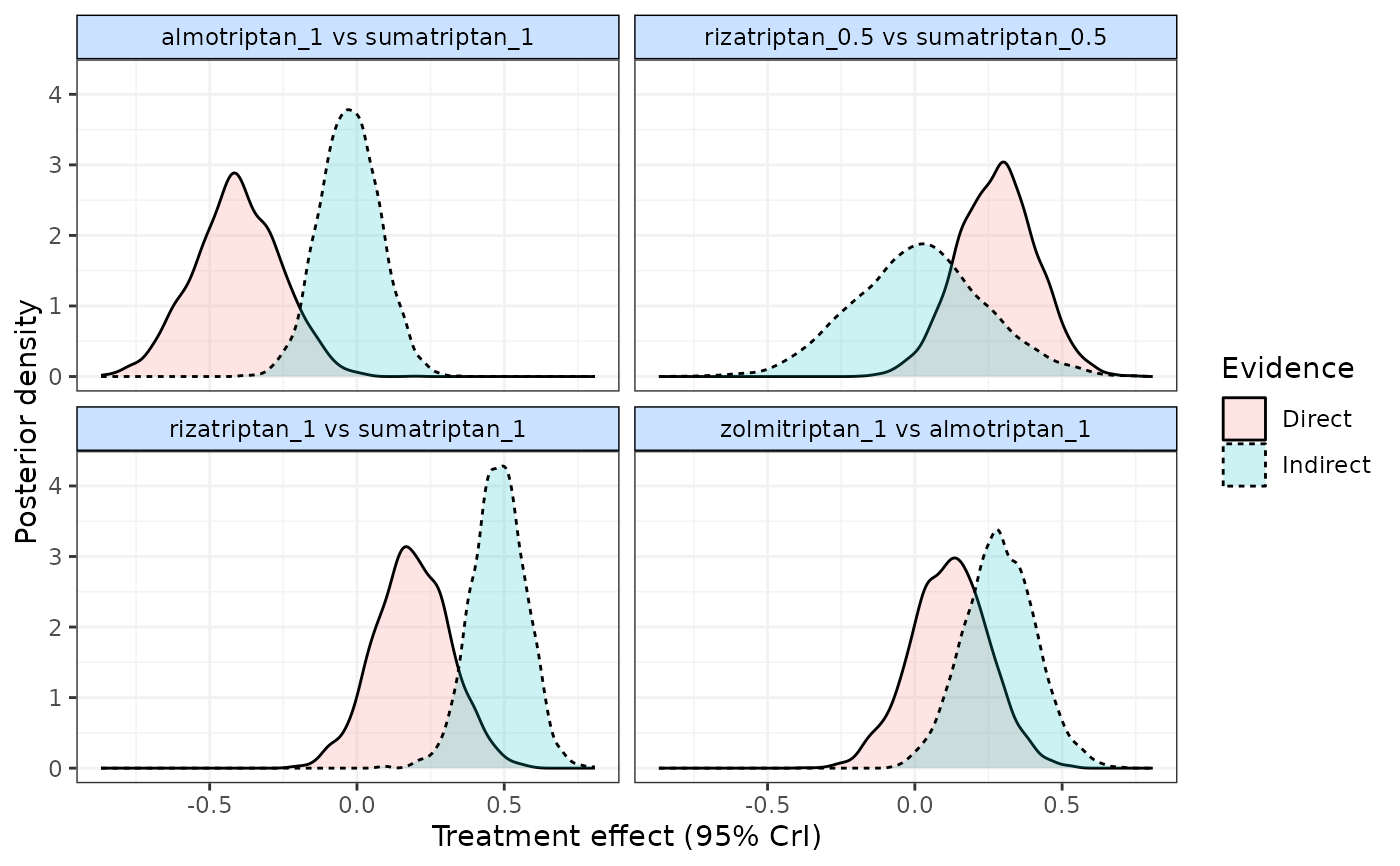 plot(split, plot.type="forest") # Plot forest plots of direct and indirect evidence
# Print and summarise results
print(split)
#> ========================================
#> Node-splitting analysis of inconsistency
#> ========================================
#> |Comparison | p-value| Median| 2.5%| 97.5%|
#> |:----------------------------------|-------:|------:|------:|------:|
#> |zolmitriptan_1 vs almotriptan_1 | 0.513| | | |
#> |-> direct | | 0.119| -0.142| 0.369|
#> |-> indirect | | 0.285| 0.051| 0.526|
#> |-> MBNMA | | 0.206| 0.041| 0.390|
#> | | | | | |
#> |rizatriptan_1 vs sumatriptan_1 | 0.171| | | |
#> |-> direct | | 0.185| -0.048| 0.430|
#> |-> indirect | | 0.482| 0.300| 0.657|
#> |-> MBNMA | | 0.389| 0.242| 0.530|
#> | | | | | |
#> |almotriptan_1 vs sumatriptan_1 | 0.127| | | |
#> |-> direct | | -0.404| -0.691| -0.119|
#> |-> indirect | | -0.024| -0.217| 0.184|
#> |-> MBNMA | | -0.148| -0.314| 0.012|
#> | | | | | |
#> |rizatriptan_0.5 vs sumatriptan_0.5 | 0.445| | | |
#> |-> direct | | 0.284| 0.028| 0.534|
#> |-> indirect | | 0.020| -0.419| 0.469|
#> |-> MBNMA | | 0.213| -0.015| 0.435|
#> | | | | | |
summary(split) # Generate a data frame of summary results
#> Comparison Evidence Median 2.5% 97.5% p.value
#> 1 zolmitriptan_1 vs almotriptan_1 Direct 0.119 -0.142 0.369 0.513
#> 2 zolmitriptan_1 vs almotriptan_1 Indirect 0.285 0.051 0.526 0.513
#> 3 zolmitriptan_1 vs almotriptan_1 NMA 0.206 0.041 0.390 0.513
#> 4 rizatriptan_1 vs sumatriptan_1 Direct 0.185 -0.048 0.430 0.171
#> 5 rizatriptan_1 vs sumatriptan_1 Indirect 0.482 0.300 0.657 0.171
#> 6 rizatriptan_1 vs sumatriptan_1 NMA 0.389 0.242 0.530 0.171
#> 7 almotriptan_1 vs sumatriptan_1 Direct -0.404 -0.691 -0.119 0.127
#> 8 almotriptan_1 vs sumatriptan_1 Indirect -0.024 -0.217 0.184 0.127
#> 9 almotriptan_1 vs sumatriptan_1 NMA -0.148 -0.314 0.012 0.127
#> 10 rizatriptan_0.5 vs sumatriptan_0.5 Direct 0.284 0.028 0.534 0.445
#> 11 rizatriptan_0.5 vs sumatriptan_0.5 Indirect 0.020 -0.419 0.469 0.445
#> 12 rizatriptan_0.5 vs sumatriptan_0.5 NMA 0.213 -0.015 0.435 0.445
# }
plot(split, plot.type="forest") # Plot forest plots of direct and indirect evidence
# Print and summarise results
print(split)
#> ========================================
#> Node-splitting analysis of inconsistency
#> ========================================
#> |Comparison | p-value| Median| 2.5%| 97.5%|
#> |:----------------------------------|-------:|------:|------:|------:|
#> |zolmitriptan_1 vs almotriptan_1 | 0.513| | | |
#> |-> direct | | 0.119| -0.142| 0.369|
#> |-> indirect | | 0.285| 0.051| 0.526|
#> |-> MBNMA | | 0.206| 0.041| 0.390|
#> | | | | | |
#> |rizatriptan_1 vs sumatriptan_1 | 0.171| | | |
#> |-> direct | | 0.185| -0.048| 0.430|
#> |-> indirect | | 0.482| 0.300| 0.657|
#> |-> MBNMA | | 0.389| 0.242| 0.530|
#> | | | | | |
#> |almotriptan_1 vs sumatriptan_1 | 0.127| | | |
#> |-> direct | | -0.404| -0.691| -0.119|
#> |-> indirect | | -0.024| -0.217| 0.184|
#> |-> MBNMA | | -0.148| -0.314| 0.012|
#> | | | | | |
#> |rizatriptan_0.5 vs sumatriptan_0.5 | 0.445| | | |
#> |-> direct | | 0.284| 0.028| 0.534|
#> |-> indirect | | 0.020| -0.419| 0.469|
#> |-> MBNMA | | 0.213| -0.015| 0.435|
#> | | | | | |
summary(split) # Generate a data frame of summary results
#> Comparison Evidence Median 2.5% 97.5% p.value
#> 1 zolmitriptan_1 vs almotriptan_1 Direct 0.119 -0.142 0.369 0.513
#> 2 zolmitriptan_1 vs almotriptan_1 Indirect 0.285 0.051 0.526 0.513
#> 3 zolmitriptan_1 vs almotriptan_1 NMA 0.206 0.041 0.390 0.513
#> 4 rizatriptan_1 vs sumatriptan_1 Direct 0.185 -0.048 0.430 0.171
#> 5 rizatriptan_1 vs sumatriptan_1 Indirect 0.482 0.300 0.657 0.171
#> 6 rizatriptan_1 vs sumatriptan_1 NMA 0.389 0.242 0.530 0.171
#> 7 almotriptan_1 vs sumatriptan_1 Direct -0.404 -0.691 -0.119 0.127
#> 8 almotriptan_1 vs sumatriptan_1 Indirect -0.024 -0.217 0.184 0.127
#> 9 almotriptan_1 vs sumatriptan_1 NMA -0.148 -0.314 0.012 0.127
#> 10 rizatriptan_0.5 vs sumatriptan_0.5 Direct 0.284 0.028 0.534 0.445
#> 11 rizatriptan_0.5 vs sumatriptan_0.5 Indirect 0.020 -0.419 0.469 0.445
#> 12 rizatriptan_0.5 vs sumatriptan_0.5 NMA 0.213 -0.015 0.435 0.445
# }